Working With Drug Data from the ChEMBL Database
When working on drug discovery projects, it's handy to have access to a set of chemical structures and associated data for marketed drugs. If you're considering introducing new functionality, someone invariably asks whether that functionality has been used in a marketed drug. It's also helpful to compare the properties of a new compound or compounds to those of marketed drugs. Early in my career, I remember a new medicinal chemist asking Josh Boger, the founder of Vertex Pharmaceuticals, what they should do on their first day of work. Boger responded, "read the Merck Index so you can see what a drug is supposed to look like". Recently a few papers have been published showing how the properties of drugs have changed over time. I thought it might be helpful to create a notebook showing how to extract and clean drug data from ChEMBL and use it for subsequent analysis.
The Jupyter notebook is available here on GitHub and can also be run here on Google Colab. In the notebook, we go through the following steps.
- Download the ChEMBL database.
- Query ChEMBL for drug data.
- Remove duplicates from the ChEMBL data.
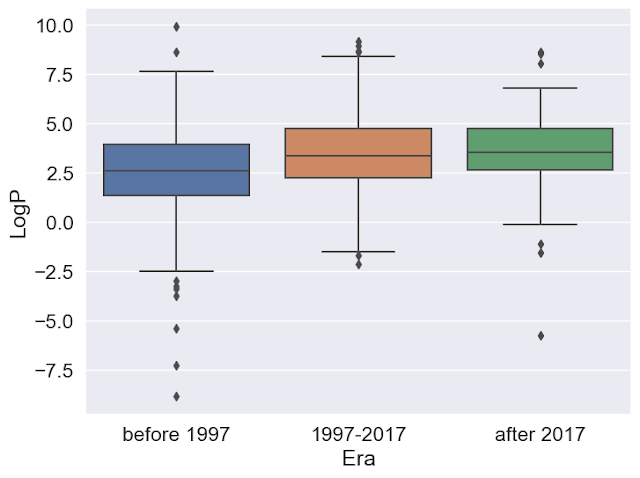
- Divide the ChEMBL data into three groups based on the first approval date, before 1997, 1997-2017, and after 2017.
- Compare the molecular weight and calculated logP distributions for the three groups and determine if the differences between groups are statistically significant.
Ultimately, we generate boxplots comparing calculated drug properties over time and heatmaps that indicate whether the differences are statistically significant. There are also a few data analysis tricks and tips that some might find helpful. I hope others will learn from this and use this notebook as a foundation for their work.



Hi Pat, thank you so very much for the wonderful tutorial and github link.
ReplyDelete1) Are there other databases you would recommend linking in for the market analytics piece?
2) How would you ask the inverse question. "If I develop a new mechanism of action, what indication should I puruse"?