Clustering Fragment Screening Hits With a Self-Organizing Map
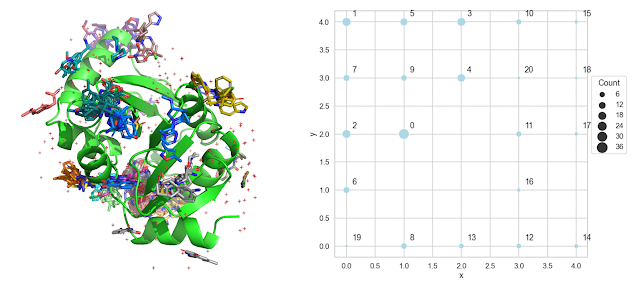
Clustering Fragment Screening Hits With a Self-Organizing Map (SOM) In a paper, " Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking ", published last year by scientists from UCSF and the Diamond Light Source, the authors reported more than 200 structures of fragments bound to the Nsp3 macrodomain of SARS-CoV-2. I wanted to dig into the supporting data from this paper and compare fragments that were binding to the same site. I couldn't find a good tool to do this, so I decided to write one. After writing the code, I thought that others might find the methodology instructive. I have a Jupyter notebook with the code on GitHub . There is also a link to run the notebook on Google Colab . Here's a quick outline of the workflow. 1. Download the structures from the PDB The supporting information for the paper has a list of the PDB ids for t...