Multiple Comparisons, Non-Parametric Statistics, and Post-Hoc Tests
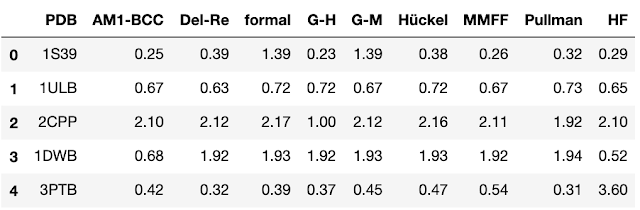
In Cheminformatics, we frequently run into situations where we want to compare more than two datasets. When comparing multiple datasets, we have a higher probability of chance correlation so we must make a few adjustments to the ways in which we compare our data. In this post, we will examine the impact of multiple comparisons and talk about methods known as post-hoc tests that can be used to correct p-values when multiple comparisons are performed. We will also make a brief foray into non-parametric statistics, a technique that is appropriate for dealing with the skewed data distributions that we often encounter in drug discovery. As usual, all of the data and the code used to perform the analyses in this post is available on GitHub . I always find it easier to understand a method when I’m presented with an example. For this case, we’ll look at a 2011 paper from Li and coworkers . In this paper, the authors compared the impact...
