
“Pay no attention to the man behind the curtain” - The Wizard of Oz
Introduction
Recently, a few groups have proposed general-purpose large language models (LLMs) like ChatGPT, Claude, and Gemini as tools for generating molecules. This idea is appealing because it doesn't require specialized software or domain-specific model training. One can provide the LLM with a relatively simple prompt like the one below, and it will respond with a list of SMILES strings.
You are a skilled medicinal chemist. Generate SMILES strings for 100 analogs of the molecule represented by the SMILES CCOC(=O)N1CCC(CC1)N2CCC(CC2)C(=O)N. You can modify both the core and the substituents. Return only the SMILES as a Python list. Don’t put in line breaks. Don't put the prompt into the reply.
However, when analyzing molecules created by general-purpose LLMs, I'm reminded of my undergraduate Chemistry days. My roommates, who majored in liberal arts, would often assemble random pieces from my molecular model set and ask, "What is this?" My usual response was, "Something that could never exist." While there was a slight chance that my roommates, with no chemistry knowledge, could have accidentally created the structure of a blockbuster drug, it was highly improbable. Examining molecules produced by LLMs gives me a similar impression. It's like someone with no chemistry background is trying to generate a molecule. As we'll explore below, the LLM-generated molecules frequently exhibit "tricks" rather than genuine insights into the underlying chemical principles.
Testing Claude’s Molecule Generation Ability
To test the molecule generation ability of Claude, a general-purpose LLM from Anthropic, I prompted it to generate analogs for hits from fragment screens. These fragment hits are small molecules with between 11 and 21 heavy atoms. As a set of test inputs, I chose 37 molecules published in the excellent “Fragment to Lead '' series that has appeared annually in The Journal of Medicinal Chemistry (JMC) since 2017. This set of JMC reviews highlights interesting papers where groups have optimized hits from fragment screens into lead molecules for drug discovery programs. Conveniently, the Supporting Information for the most recent review has a spreadsheet with the SMILES for hit/lead pairs from all the previous editions. For each of the 37 fragment SMILES, I used the Anthropic Python API to provide Claude with the prompt above and examined the structures of the molecules generated by the LLM.
Previously, numerous authors have compared the chemical space covered by analogs produced by various generative models, including general-purpose LLMs. They typically calculate the molecules' fingerprints, then project them into two dimensions using a technique like t-Distributed Stochastic Neighbor Embedding (tSNE). The tSNE plot provides a convenient means of examining where the generated molecule sets overlap. Rather than trying to obtain a quantitative measure of chemical space coverage, I wanted to see if the analogs produced by Claude made sense from a medicinal chemistry perspective. When performing SAR exploration, we want to design a set of analogs that will enable us to understand activity drivers and determine which molecules to make next. I tried to assess whether Claude was generating analogs that would allow me to explore and understand SAR. I freely admit that this is a qualitative analysis influenced by my biases. Moreover, this small sample may only partially reflect some LLM use cases.
String Manipulation Masquerading as Chemistry
Looking through the Claude output, it quickly became apparent that the LLM frequently uses a few “tricks” to generate analogs. The first is to remove the last token from a SMILES and replace it with common functional groups. Consider the SMILES below.
Nc1cc(c[nH]c1=O)C(F)(F)F
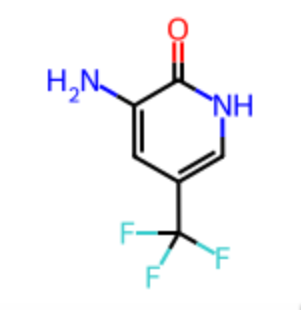
Claude removed the fluorine from the end of the SMILES and replaced it with several halogen alternatives.
Nc1cc(c[nH]c1=O)C(F)(F)Cl
Nc1cc(c[nH]c1=O)C(F)(F)Br
Nc1cc(c[nH]c1=O)C(F)(F)I
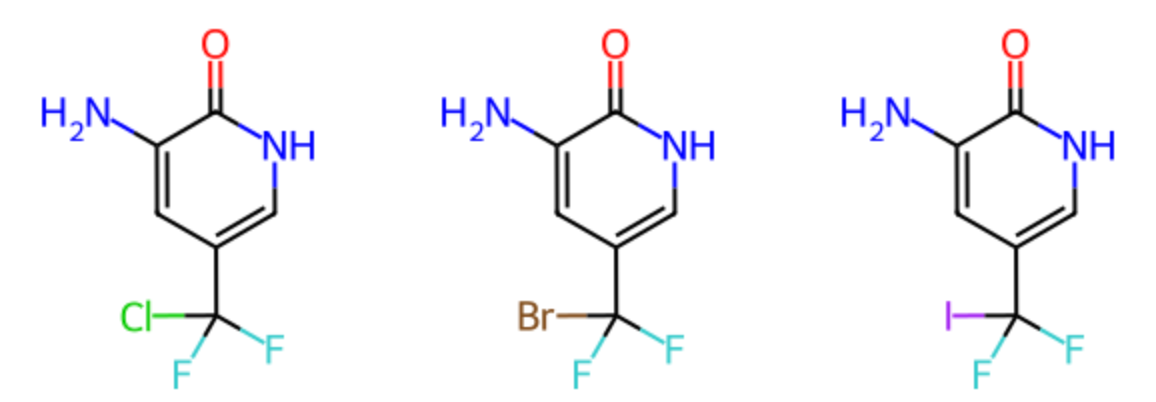
Claude also further truncated the SMILES to add additional functional groups. In the example below, two fluorines were removed and replaced by two chlorines.
Nc1cc(c[nH]c1=O)C(F)(Cl)Cl
While the analogs generated above are valid, the resulting exploration is somewhat limited. Unfortunately, Claude doesn’t understand SMILES, and it attempts to append invalid strings like “CF3” and “CH3,” resulting in invalid SMILES that can’t be parsed. In my experiments, approximately 20% of the generated structures contained invalid characters and could not be parsed.
Nc1cc(c[nH]c1=O)C(F)(F)CF3
Nc1cc(c[nH]c1=O)C(F)(F)CH3
Another common strategy that Claude employs is to find a methyl group in parentheses in a SMILES string and replace the methyl “C” with another functional group. For example, consider the SMILES below, where a methyl in parentheses is shown in bold.
CN1C[C@@H](O)[C@H](C1=O)c2ccc(C)cc2
As mentioned above, it’s straightforward for Claude to generate new molecules by simply making string substitutions.
CN1C[C@@H](O)[C@H](C1=O)c2ccc(C=C)cc2
CN1C[C@@H](O)[C@H](C1=O)c2ccc(C#C)cc2
CN1C[C@@H](O)[C@H](C1=O)c2ccc(CCl)cc2
CN1C[C@@H](O)[C@H](C1=O)c2ccc(CBr)cc2
CN1C[C@@H](O)[C@H](C1=O)c2ccc(CI)cc2
CN1C[C@@H](O)[C@H](C1=O)c2ccc(CF)cc2
CN1C[C@@H](O)[C@H](C1=O)c2ccc(CCF)cc2
CN1C[C@@H](O)[C@H](C1=O)c2ccc(CCI)cc2
It’s also possible for Claude to do some genuinely silly things. In one case I examined, Claude repeatedly substituted a longer and longer alkyl chain for a methyl group in parentheses.
CN1C[C@@H](O)[C@H](C1=O)c2ccc(CCCC)cc2
CN1C[C@@H](O)[C@H](C1=O)c2ccc(CCCCC)cc2
CN1C[C@@H](O)[C@H](C1=O)c2ccc(CCCCCC)cc2
CN1C[C@@H](O)[C@H](C1=O)c2ccc(CCCCCCC)cc2
CN1C[C@@H](O)[C@H](C1=O)c2ccc(CCCCCCCC)cc2
CN1C[C@@H](O)[C@H](C1=O)c2ccc(CCCCCCCCC)cc2
CN1C[C@@H](O)[C@H](C1=O)c2ccc(CCCCCCCCCC)cc2
CN1C[C@@H](O)[C@H](C1=O)c2ccc(CCCCCCCCCCC)cc2
CN1C[C@@H](O)[C@H](C1=O)c2ccc(CCCCCCCCCCCC)cc2
CN1C[C@@H](O)[C@H](C1=O)c2ccc(CCCCCCCCCCCCC)cc2
CN1C[C@@H](O)[C@H](C1=O)c2ccc(CCCCCCCCCCCCCC)cc2
CN1C[C@@H](O)[C@H](C1=O)c2ccc(CCCCCCCCCCCCCCC)cc2
CN1C[C@@H](O)[C@H](C1=O)c2ccc(CCCCCCCCCCCCCCCC)cc2
CN1C[C@@H](O)[C@H](C1=O)c2ccc(CCCCCCCCCCCCCCCCC)cc2
CN1C[C@@H](O)[C@H](C1=O)c2ccc(CCCCCCCCCCCCCCCCCC)cc2
CN1C[C@@H](O)[C@H](C1=O)c2ccc(CCCCCCCCCCCCCCCCCCC)cc2
CN1C[C@@H](O)[C@H](C1=O)c2ccc(CCCCCCCCCCCCCCCCCCCC)cc2
Here’s a pictorial view of what the LLM is doing for those less familiar with SMILES strings. This epitomizes the old medicinal chemistry adage “methyl, ethyl, futile,” describing pointless analogging.
While capable of producing generally valid SMILES, the methods above fall short in their utility for SAR exploration. In my analysis of examples, most substitutions occurred at a singular position on the scaffold. This approach is less effective when embarking on initial SAR exploration.
Sometimes, Claude adopts what appears to be a medicinal chemist's approach. In the example below, the LLM seems to perform a "nitrogen walk," generating a sequence of pyridine and pyrimidine isomers to investigate the aromatic ring on the left side of the molecule. The number below each structure represents the frequency of the ring system's appearance in the ChEMBL database. The LLM uses common ring systems, and the structures below demonstrate a logical strategy for SAR exploration.
Unfortunately, some other analogs Claude generated for the same molecule demonstrate its lack of chemistry knowledge. The ring systems on the left side of the molecules below don’t appear in ChEBML and are probably unstable.

I could go on all day with additional examples of silly molecules that Claude generated, but I’m probably reaching the limit of the reader’s patience. The point I want to make is that a general-purpose LLM doesn’t understand Chemistry and can’t be expected to provide reasonable suggestions. One could argue that the poor performance of the LLM is due to my lack of “prompt engineering” skills. Perhaps if I had been able to divine the perfect prompt, I might have achieved a better result. However, rather than fumbling and randomly attempting to define the ideal prompt, I’d prefer to employ a more appropriate tool.
There Are Better Ways To Do This
Using a general-purpose LLM as a molecule generator, in many ways, feels like using a butter knife as a screwdriver. Yes, it (sort of) works, but there is a better way. Over the last few years, dozens of valid and valuable approaches to analog generation have been published and are available as open-source software packages. While installing and using some of these packages can be challenging, I’ve found it worthwhile.
I divide these generative approaches into three categories. First is what I refer to as “knowledge-based replacements.” This approach can be described by paraphrasing the amazon.com slogan “people who bought this also bought …” as “chemists who made this also made …”. Using techniques like
matched molecular pairs and
matched molecular series, one can search databases like
ChEMBL for pairs of molecules where one functional group was used to replace another. By tabulating these changes and the associated atom environments in a database, one can build a tool that generates reasonable analogs of an input molecule. Open-source tools like
CReM and
MMPDB are prime examples of this "knowledge-based replacements" approach.
Recent applications of neural networks have led to the emergence of two additional categories of generative molecular design: latent space models and recurrent neural networks (RNNs). Inspired by earlier work on image generation, latent space models comprise two components: an encoder that translates a chemical structure into a low-dimensional vector representation and a decoder that can reconstruct the chemical structure from this low-dimensional representation. By randomly perturbing the latent space vector and decoding the perturbed vector, we can generate the structures of related molecules. Several variations of latent space models have emerged in recent years, including generative adversarial networks (GANs) and variational autoencoders (VAEs). Among these, MolGAN and COATI stand out as open-source implementations.
The final category of molecule generators comprises RNNs and associated techniques like transformers. These methods, stemming from text generation algorithms, are designed to learn probability distributions for tokens (usually characters) that follow other tokens in a character string. These algorithms underpin the suggestion features in widely used email and text messaging applications. For instance, when I type "would you like to meet for" in a text message, there's a high probability the next word will be "coffee" or "lunch." Similarly, an RNN trained on a sizable collection of SMILES strings can learn that the pattern "c1ccccc1C(=O)" is likely to be followed by "C" or "O." By sampling from these distributions, an RNN can generate mostly reasonable structures. The REINVENT program from AstraZeneca is probably the most widely used open-source RNN molecule generator. Morgan Thomas’ GitHub repository is also an excellent resource for RNN related methods.
The methods discussed in this section differ from general-purpose LLMs, like Claude, in that they have been specifically trained on chemical data. The training data typically comprises millions of molecules from the ChEMBL, PubChem, or ZINC databases. In contrast, general-purpose LLMs are inexact search engines trained on a vast range of content from the internet. As Gary Marcus succinctly put it, "LLMs don't learn concepts; they learn similarity to the training corpus." Given the availability of numerous domain-specific tools, why would anyone opt to use a general-purpose LLM for molecule generation?
Some Parting Thoughts on LLMs for Science
Over the last year, there has been tremendous hype around applying LLMs in science. One aspect that could be more explicit in this discussion is the difference between looking something up and understanding a concept. Many papers simply look at an LLM's ability to answer things like exam questions. However, we should also attempt to determine whether the LLM understands the subject matter or simply looked up a similar question in its training corpus.
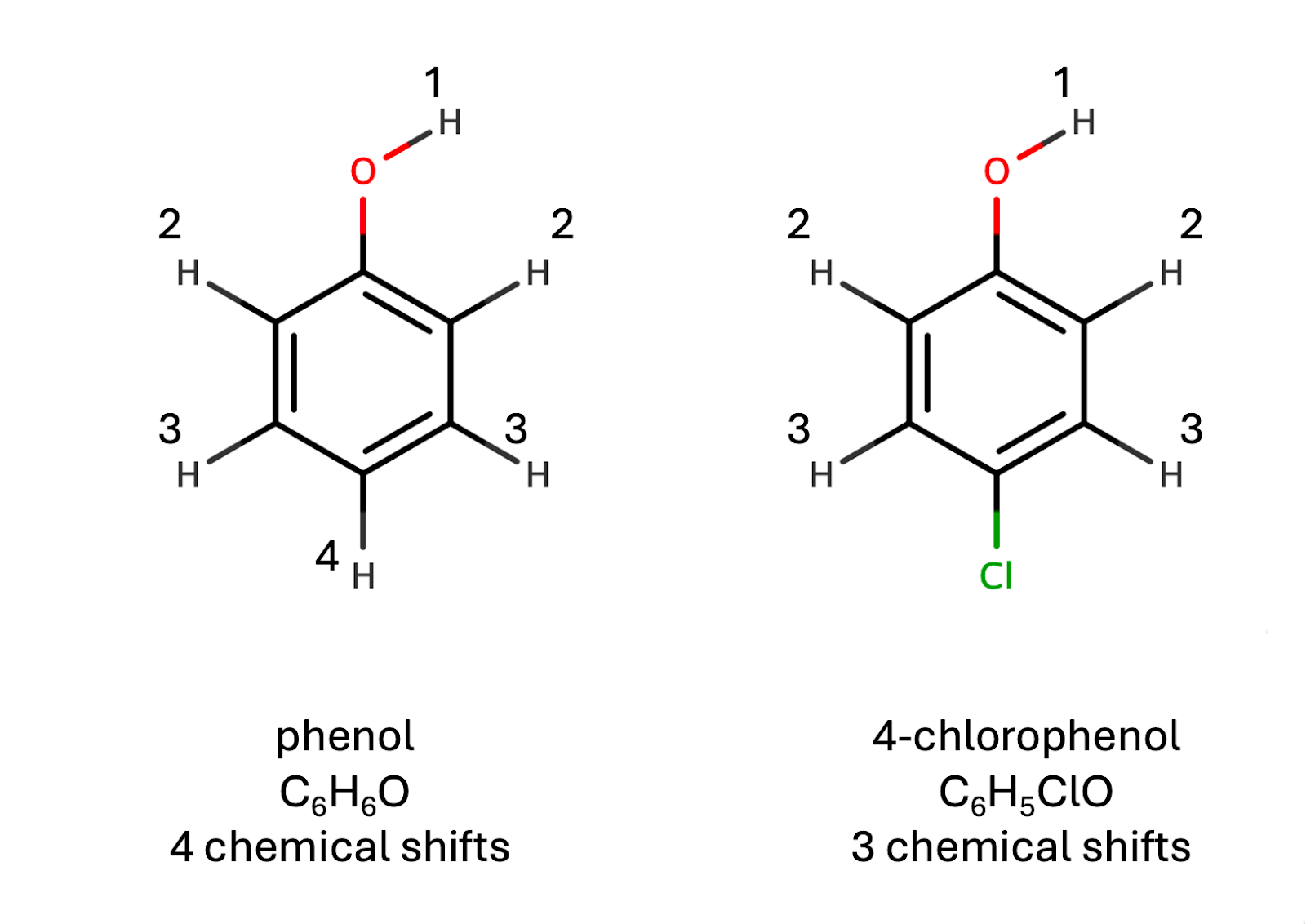
As an illustrative example, consider predicting the number of distinct proton NMR chemical shifts in a molecule. This problem commonly appears in homework assignments and exams for introductory organic chemistry students. Predicting the number of distinct chemical shifts requires understanding symmetry and molecular geometry to determine the number of unique hydrogen chemical environments within a molecule. To illustrate this concept, we will examine two simple molecules: phenol and 4-chlorophenol (depicted in the figure below). In the figure, each number represents a distinct chemical environment. We can see that phenol has four distinct hydrogen environments, while 4-chlorophenol has three.
Here’s a prompt and a response from ChatGPT.
Prompt: How many distinct chemical shifts does the proton NMR spectrum of phenol have?
Response: The proton NMR spectrum of phenol has four distinct chemical shifts. This is because the six hydrogen atoms (protons) of phenol occupy four different chemical environments.
Pretty impressive, right? When ChatGPT produces this response, it gives the illusion of understanding chemistry. However, when we look at the helpful reference provided by ChatGPT, we see that this information was derived from a web page with an extensive analysis of the NMR spectrum of phenol.
If we make a small change to the prompt, we get another very confident but completely incorrect response. In fact, the response doesn’t even contain the correct number of hydrogen atoms.
Prompt: How many distinct chemical shifts does the proton NMR spectrum of 4-chlorophenol have?
Response: The proton NMR spectrum of 4-chlorophenol has five distinct chemical shifts. This is because the six hydrogen atoms (protons) of 4-chlorophenol occupy five different chemical environments.
ChatGPT references the same web page referenced above. It has effectively copy-pasted the same response.
Don’t get me wrong—there’s nothing wrong with a tool that correctly looks things up. I frequently use LLMs to point me to the appropriate literature and help me answer scientific questions. However, as these methods become more pervasive, it’s essential to clearly understand what they can and can’t do.
An Added Note on GPT4-io
When I was finishing this post, OpenAI released its latest version, GPT4-o1. Of course, I had to try the questions above with the new version. For the phenol NMR question, GPT4-o1 again gave the correct answer with a more expansive explanation. For 4-chlorophenol, the latest version provides the correct answer; perhaps things are improving. I also tried a few analog generation examples, which, on a qualitative level, seemed more interesting than the analogs created by Claude. GPT4-o1 was changing both the scaffold and the substituents. However, there were also plenty of silly analogs. One of the most ridiculous is shown below.
Where Do We Go From Here
In 2023, Chris Murphy, US Senator from Connecticut, posted the tweet below on Twitter (I refuse to call it X).
Of course, this is ridiculous. ChatGPT doesn't "teach itself" anything and doesn't understand basic, let alone advanced, Chemistry. LLMs for science are still in their infancy, and the field is progressing rapidly. As the field progresses, we need better ways to test an LLM's ability to capture concepts rather than simply examining its ability to respond to exam questions where the answers can be looked up.
Acknowledgments
I'd like to thank Santosh Adhikari for helpful discussions on an early version of this post.




Hi Pat, thank you for this post! A few questions/notes:
ReplyDelete1) It is known that LLMs perform better when the generation for the same prompt is repeated multiple times. For the analog generation, a workflow similar to the one described in https://arxiv.org/abs/2406.07394 might be implemented, by repeating the generation of SMILES via one prompt and then checking their validity with another prompt. Did you check if the quality of the generated SMILES improves when you run it multiple times?
2) I used the same prompt with the new o1 model from OpenAI. Although it is claimed to have better quality of reasoning than 4o/Claude, the results were quite similar to what you observed with Claude. O1 was even worse with stereochemistry, as it forgot to put square brackets around the stereocenters, so all the SMILES generated were invalid.
It did better with the first structure you mentioned, having the CF3 group. 88 out of 106 SMILES were valid. For some reason, only the CF3 group was modified, and no changes were made in the six-membered ring. o1 really preferred amide group as a replacement for fluorine. Many of the representatives had systematic variations, such as replacing F with other halogens or the nitrile group. There were some chemistry-concise examples, such as changing F to the morpholine amide (NC1=CC(C(C(N2CCOCC2)=O)(F)F)=CNC1=O), but also a bunch of really atrocious ones, like having N-I bonds or replacing each fluorine with an oxime. I also didn't observe the addition of long alkyl chains.
Overall, in its current state, o1 is not useful for such tasks.
I think this sums it up perfectly: "Given the availability of numerous domain-specific tools, why would anyone opt to use a general-purpose LLM for molecule generation?". Also, coming up with new ideas does not seem to be the bottleneck of drug design. Minor correction: I think the t in tSNE stands for t-distributed
ReplyDelete