Filtering Chemical Libraries
This post was partially motivated by a recent post from Karl Leswing describing how to use the DeepChem package to do virtual screening on a large database. As part of the tutorial, Karl used the HIV sample file that is part of the DeepChem distribution to build a model. This model was then used to select compounds from the ZINC database. The tutorial is nice and the methodology is explained in a manner that is easy to follow. The problem is that the molecules selected by the model are not what I would consider "drug-like".
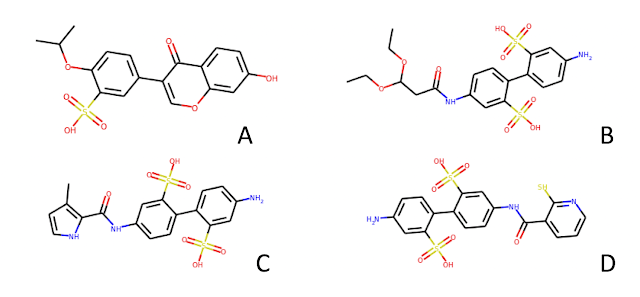
The molecules reported in Karl's post have aryl sulfonic acid groups. In fact, molecule D has 2 sulfonic acids AND a thiol. To someone who has spent a bit of time looking through HTS data, these molecules scream false positive. If an enterprising young computational chemist were to take this set of hits to an experienced medicinal chemistry colleague, my guess is that he/she would simply shake his/her head and say something like "that's nice, please come back when you have something real".
This brings us to the topic of this post, filtering chemical libraries. Over the last 25 years, many people in the cheminformatics community (including me), have put together sets of substructure queries designed to identify compounds which are likely to produce artifacts in biochemical or cellular assays. In the late 1990s, Mark Murcko and I put together a set of filters that we called REOS (Rapid Elimination of Swill). This wasn't the first and certainly wasn't the last such effort. Some examples from the literature include work by groups at
- Amgen
- University of New Mexico
- Lilly
- Perhaps the most notable (some might say notorious) is the PAINS filters developed by Bael and Holloway.
The controversy came to a boil with a 2017 paper by Tropsha and coworkers that questioned recent guidelines on the use of PAINS filters by the editors of the Journal of Medicinal Chemistry (JMC). In the interest of full disclosure, I am a member of the JMC Editorial Advisory Board and helped to draft those guidelines. It should be noted that the JMC guidelines do not state that a paper will be rejected solely based on the presence of PAINS substructures. The guidelines simply state that screening hits containing PAINS substructures should be supported by additional experimental evidence (e.g. SAR, structural information, orthogonal assays etc.). In truth, any paper reporting hits from virtual screening should demonstrate the purity and the validity of the hits. Tropsha argued that the PAINS filters are somewhat arbitrary and have limited experimental validation. He also pointed out that a number of FDA approved drugs contain PAINS filters. The editors of JMC and a number of related ACS journals responded with a detailed editorial supporting the original guidelines.
There is a lot more I could say about structural alerts, but that's enough of a preamble. After all, this blog is called Practical Cheminformatics, so let's get to it. I'm sure that many people may want to run a set of in-silico filters on their compounds, but simply don't know where to start. Fear not, you've come to the right place.
The ChEMBL database now contains a database table that features a large collection of substructure filters. The table is called "structural_alerts" and, in ChEMBL 23, contains more than a thousand structural alerts from 8 different alert sets. Of course, this may not be a lot of use to you if you're not a database geek and a cheminformatics geek. Again, fear not, I've pulled the structural alerts from the database, cleaned up a few that misbehaved, and wrapped this up in an easy to use Python script.
The script, rd_filters.py, is available on my GitHub site. The GitHub site has some examples, but let's take a look at what happens if we run one of the ChEMBL structural filter sets on the HIV dataset from DeepChem. The file is deepchem/examples/hiv/HIV.csv. First off, rd_filters.py expects a SMILES file and HIV.csv is a csv file. No worries, we can fix this.
awk -F',' 'NR > 1{printf("%s MOL%04d\n",$1,NR-1)}'
In case you're not an awk geek, this uses a comma as a delimiter, skips the first line (which is a header), prints the first token on the line (the SMILES string), prints MOL followed by the line number padded with 4 zeros. I subtract 1 from the line number (NR) because we skipped the first line. Note that the file HIV.smi is also on the GitHub site.
Ok, now that we have a SMILES file, the fun can begin. If we want to run HIV.smi with the default filters and the default Inpharmatica structural alerts, we will get output that looks like this.
rd_filters.py filter --in HIV.smi --prefix out
using 8 cores
Using alerts from Inpharmatica
Wrote SMILES for molecules passing filters to out.smi
Wrote detailed data to out.csv
18499 of 41913 passed filters 44.1%
Elapsed time 22.87 seconds
There are a few things to note here.
- First off, the script runs in parallel across multiple cores. For the Python geeks in the crowd, this is due to the magic that is Pool.map. By default, it uses all of the available cores, but this can be controlled with the --np flag (see the README.md for more information). It's pretty fast, 22 seconds for 42K compounds on my MacBook Pro.
- We used the structural alerts from the Inpharmatica set in the ChEMBL table, we can modify the configuration file rules.json to change to a different set of structural alerts.
- The script outputs 2 files, one with the SMILES for the compounds which pass all of the alerts and property filters, another with detailed filtering results.
- Only 44% of the compounds in the input file passed the filters. This is not good.
awk -F',' '{print $4}' out.csv | sort | uniq -c | sort -rn | head
23450 OK
2643 Filter9_metal > 0
2401 Filter44_michael_acceptor2 > 0
1760 Filter82_pyridinium > 0
1296 Filter14_thio_oxopyrylium_salt > 0
1086 Filter20_hydrazine > 0
930 Filter39_imine > 0
894 Filter41_12_dicarbonyl > 0
697 Filter5_azo > 0
614 Filter1_2_halo_ether > 0
6% of the compounds were eliminated by a single rule "Filter9_metal", let's look at the SMARTS for this alert.
[!#1!#6!#7!#8!#9!#15!#16!#17]~[*,#1]
This filter is designed to identify compounds with atom types outside those typically found in organic molecules
H,C,N,O,F,P,S,Cl. As you can see in the figure below, this filter does indeed pick up a lot of silly molecules that probably shouldn't be in anyone's screening collection.
* Copper complexes
* Zinc complexes
* Boronates
* Cobalt complexes
* Arsenates
Of course, the astute reader will also note that the filter tosses compounds with bromine, something I wouldn't do (the disulfide in MOL0049 is another story). This highlights the fact that these sorts of things can't just be used blindly, you have to run the filters, take a look at what is being eliminated and adjust as necessary.
To sum all of this up.
- Most screening collections contain a lot of molecules that probably shouldn't be there.
- Structural alerts provide a means of identifying and potentially eliminating some of these potentially problematic molecules.
- The rd_filters.py script provides a convenient way of applying a number of different sets of structural alerts to a set of molecules. Please give it a try.
- A lot of these filters are somewhat subjective and based on people's experience. 4 of the 8 rule sets in ChEMBL will reject the sulfonic acids shown at the beginning of this post, the other four will not.
Another thing we haven't talked about is, which of the 8 ChEMBL structural alert sets is the best one to use. That's a discussion for another day.

This work is licensed under a Creative Commons Attribution 4.0 International License.



Comments
Post a Comment